Research areas
Species genetic variability strongly affects their resilience in the face of rapid and drastic changes. Aquatic and terrestrial organisms are confronted with many challenges, many of them human-made, such as temperature changes, habitat availability, eutrophication and shifts in biotic interactions such as predation. Given the importance of freshwater resources to human populations, a better understanding of ecosystem functioning is crucial for guiding conservation and protection efforts for these habitats.
The research group is focussing on water fleas and takes advantage of two reproduction characteristics of this group: the resting stages produced through sexual reproduction, that constitute biological archives and can be hatched decades later; and the clonal reproduction mode that allows life-history experiments and RNA analyses to be conducted to disentangle genetic from environmental effects. My research integrates a variety of disciplines, such as population genetics, bioinformatics, experimental biology, freshwater ecology, genomics, and transcriptomics.
Recombination and Hybridization in a Daphnia species complex
The Cordellier Lab substantially contributed to our current knowledge of Daphnia genetics, by sequencing and assembling a high-quality genome assembly and inferring mutation rate for Daphnia galeata, and assessing genetic variability of the species complex. Sustained gene flow seems to occur in the DLSC. However, interspecific differentiation is highly variable across the genome, and the underlying mechanisms are unclear. How are selection and recombination shaping introgression? To address this question, we are planning to sequence and assemble the genomes for two other species in the species complex, an essential step to address many of the planned projects outlined here. We will also estimate species specific recombination rates using bulk gamete sequencing.
Population genomics through time with resting egg banks
Many lakes in Northern Germany are inhabited by one of more species of the DLSC which are hybridizing, equating to a naturally replicated hybridization “experiment”. What’s more, the Daphnia resting eggs accumulate in the sediment and constitute a biological record of genetic changes past. By analysing these records in the light of data gathered in the project outlined above, we want to test whether the combination of selection and recombination indeed leads to predictable introgression patterns. After sampling sediment cores in select lakes, we will use isotope dating and analyse resting egg density. Genomic data will be obtained from single resting eggs after whole genome amplification, and hybridization patterns analysed with population genomics tools.
Interspecific variation in gene expression in hybridizing Daphnia species
Both food limitation and predation affect resource allocation and impact life history traits, with direct consequences on population growth and thus fitness. Therefore, genes involved in the response to predation and food quality are likely under selection and might contribute to create “islands of divergence” in the DLSC.
Are closely related species using the same gene repertoire when facing stressors such as predator cues and varying food quality/quantity? How are opposing selective forces „dealt“ with? We use gene expression analysis to answer these questions, and to identify genomic regions that might explain the maintenance of species boundaries. Our analysis encompasses gene expression profiling, network analysis, and in silico study of promoter regions.
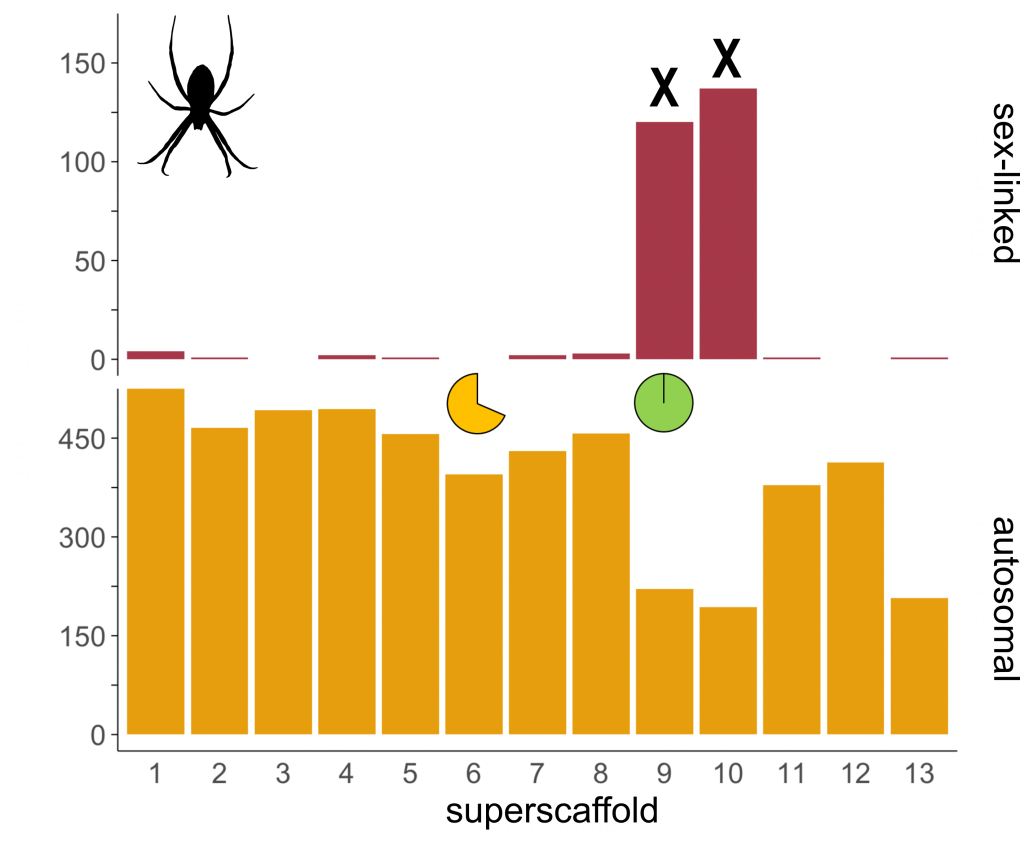
Genome and sex chromosome evolution in spiders
Many spider species exhibit highly biased sex ratios, and present striking examples of sexual selection and sexual dimorphism. Sex chromosomes are thought to be key to the establishment of separate male and female phenotype. We want to build on preliminary results hinting at high sex chromosome conservation among spider species and thus contribute to our understanding of sex chromosome evolution in the X0 system by implementing the following: systematic analysis of sex chromosomes in spider lineages throughout the phylogenetic tree, gene expression analysis in both sexes in the early stages of development, to understand dosage compensation mechanisms and test the faster-X hypothesis, establishment of molecular markers to sex individuals in early stages.